Molecular Screening
Overview
The Molecular Screening Shared Resource (MSSR) provides transformative drug discovery services to the UCLA Health Jonsson Comprehensive Cancer Center research community and beyond. The MSSR offers a comprehensive array of high throughput screening (HTS) services enabling UCLA Health JCCC investigators to discover new molecular targets, find small and large molecule drugs for these targets, and characterize and optimize candidate molecules in preparation for clinical assessments.
Leadership
Contact Information
California NanoSystems Institute
570 Westwood Plaza
Bldg 114 Room 6310
Telephone: 310-794-1974
Email: [email protected]
Website: Molecular Screening Shared Resource at ucla.edu
Hours of Operation
9:00 a.m. to 5:00 p.m. PST, Mon to Fri

Services
The MSSR delivers high throughput screening to the UCLA research community. Features of the MSSR include:
High Throughput Screening (HTS) technology
The services provided include the use of high throughput screening (HTS) technology, a total of over 300,000 compounds in various libraries, arrayed genome wide sets for CRISPR, cDNA, shRNA and siRNA for mouse and/or human. We also perform data analysis and cheminformatics taking advantage of deep learning approaches using AI and neuronal networks. The MSSR has automated equipment and data collection that make it possible to test > 100,000 compounds on a system of interest per day. Any readout is addressable: From simple plate reader based luminescent reporter gene readouts all the way to highly multiplexed high content screening applications in 3D that rely on our high throughput confocal capabilities.
Chemical Libraries
The libraries available at the MSSR cover FDA approved drugs for re-purposing, chemical genomics libraries (2,700 annotated kinase inibitors, 1,200 GPCR modulators, 2,000 natural compounds etc.) for dissecting biological pathways, and various larger libraries for drug discovery. We also offer libraries of drugs in clinical trials such as kinase inhibitors. The larger the compound collection, the greater chances of finding a hit. Currently, MSSR has over 300,000 chemicals in direct supply, a total of 500,000 compounds accessible in our network and plans to expand even further.
Databases
The MSSR maintains databases of all assays run and their results. By cross referencing and collaborating with researchers who have employed related assays, additional information may be gained providing deeper insight into processes fundamental to health and disease.
The MSSR is currently funded by the UCLA Health Jonsson Comprehensive Cancer Center, the Department of Molecular and Medical Pharmacology, the Department of Microbiology, Immunology and Molecular Genetics, and the Dean's office, David Geffen School of Medicine and the California NanoSystems Institute.
Assay Development
- Breakeven Internal Rate: $3105.94
- JCCC Rate: $606.49
- External Rate (OH): $4317.26
- External Rate (OH + MU): $4317.26
Small Molecule Screening (up to 100K)
- Breakeven Internal Rate: $2704.00
- JCCC Rate: $527.67
- External Rate: $4317.26
Small Molecule Screening (additional 100K)
- Breakeven Internal Rate: $3800.86
- JCCC Rate: $742.36
- External Rate: $5283.20
CRISPR
Access to Clones (cDNA, shRNA and CRISPR)
- Breakeven Internal Rate: $59.43
- JCCC Rate: $11.58
- External Rate: $82.61
Full Genome cDNA Screening
- Breakeven Internal Rate: $2834.48
- JCCC Rate: $553.24
- External Rate: $3939.93
shRNA or CRISPR Screening (human, per sub-library)
- Breakeven Internal Rate: $2986.06
- JCCC Rate: $583.12
- External Rate: $4150.63
siRNA Screening (human, genome wide)
- Breakeven Internal Rate: $2986.07
- JCCC Rate: $583.13
- External Rate: $4150.63
siRNA Screening (mouse, genome wide)
- Breakeven Internal Rate: $ 2985.96
- JCCC Rate: $586302
- External Rate: $4150.49
Full Technical Support (per hour)
- Breakeven Internal Rate: $ 32.57
- JCCC Rate: $ –
- External Rate: $ 45.28
Data Analysis (per hour)
- Breakeven Internal Rate: $156.08
- JCCC Rate: $30.55
- External Rate: $216.95
How We Work
The MSSR works in close conjunction with each Principle Investigator. In addition to infrastructure and chemical or biological libraries the MSSR provides the necessary expertise to guide a given HTS project towards fruition. We have adopted a collaborative workmodel in which typically one member of the Principle Investigator’s laboratory is working with the MSSR team on a given project. This ensures that a person with detailed knowledge of the biology which stands behind each project is available at all times as well as high quality results and value added to the project.
Operating Policies
The MSSR provides advanced, innovative instrumentation and services for the discovery and translation of small and large molecules into clinical candidates. We are open to academic as well as industrial clients and have experience working with large pharmaceutical companies and smaller biotech companies alike.
The MSSR works in close conjunction with each Principle Investigator. In addition to infrastructure and chemical or biological libraries, the MSSR provides the necessary expertise to guide a given HTS project towards fruition. We have adopted a collaborative workmodel in which typically one member of the Principle Investigator’s laboratory is working with the MSSR team on a given project. This ensures that a person with detailed knowledge of the biology which stands behind each project is available at all times, as well as high quality results and value added to the project.
Consultations
Consultations are free, and we strongly suggest that you take advantage of the service. During this initial appointment, we'll discuss your target of interest, libraries, general screening procedures, and (if needed) assay adaptation and controls.
Scheduling
Instrument time and library screens must be scheduled at least a week in advance. This allows time to consult, adapt the assay to high throughput mode and run controls, if necessary. Scheduling a consultation appointment well in advance (two weeks or more) of the projected screen date is advised.
Canceling
Cancelations should be made at least 24 hours in advance. Chemical libraries must be thawed in order to perform a screen. Each freeze-thaw cycle has the potential to damage chemical entities in the libraries. By canceling before the libraries are thawed, we avoid unnecessary risk to the libraries. In addition, by canceling in advance, other users can schedule to use instruments and to perform screens. So, please be considerate.
Library Handling and Instrument Operation
By MSSR personnel and trained users only.
Acknowledgements/Grants
If you plan to write a grant or write a paper including the screening you plan to perform at the MSSR, please let us know. We need to keep records of these. Such records are needed when we apply for money to support the MSSR. Recharge does not cover the entire cost of running a screen. Please help us to keep the costs to users as low as possible.
Access Prioritization
The MSSR has so far been able to accommodate all requests. Should the need arise, the MSSR advisory board will resolve prioritization conflicts.
Expertise
The Director of the MSSR, Robert Damoiseaux, is an HTS expert with an extensive background in the industry. His research interests are focused on the development of novel HTS technologies, and he is committed to supporting the HTS needs of the Cancer Center and UCLA investigators by implementing these technologies and providing guidance and oversight to all projects. Damoiseaux received his Ph.D. at the University of Lausanne under Dr. Kai Johnsson (now Director of the Max Planck Institute for Medical Research, Munich) and then joined the Institute for Functional Genomics where he was in charge of the development of the next generation assay platform for proteases. He authored >150 manuscripts and patents on HTS and synthesis (h-index 57), and was recognized as an expert author in Wiley's Development of Therapeutic Agents Handbook. Damoiseaux is a member of the cancer center, the California NanoSystems Institute (CNSI) and the Crump Institute for Molecular Imaging, and was also a member of the NIH study section and special emphasis panel reviewing HTS grants and frequently reviews for the NIH HEAL initiative. The most advanced MSSR-originated molecule (TRE-515) just passed Phase I clinical trials for solid cancers and also achieved orphan drug designation Optic Neuritis. The most advanced theragnostic antibody DUNP19 that was developed with the aid of the MSSR has obtained orphan drug designation by the FDA in 2023 for osteosarcoma and is currently in production for clinical trials. Dr. Damoiseaux is a frequent co-investigator and co-PI on grants with Cancer Center members, and these grants provide additional stability to the MSSR as they provide funding for projects and associates.
Currently, the MSSR staff includes Connie Yuen, a Berkeley trained Masters Research Associate, provides hands-on expertise in assay development, screening execution, and library management, and Cameron Taylor, who is tasked with developing high throughput cell line generation capabilities and CRISPR-based screening approaches.
Equipment
Thermo Spinnaker System I
- Flex Station II: 96 and 384 well plate reader (Molecular Devices): GPCR and ion channel assays and general fluorescence applications.
- Victor 3V: 96 or 384 and 1536 well : UV-VIS absorbance, luminescence, fluorescence polarization, fluorescence, time resolved fluorescence assay.
- Biomek FX: Liquid handler for compound transfer and cherry picking. It is equipped with a custom built pin tool and transfers 384 compounds in 85 seconds.a
- Thermo Spinnaker robotic arm on a rail: delivers and transfers plates from plate hotels or incubators onto the liquid handlers, plate readers, etc.
- Liconic STX 220: CO2 plate incubator with a capacity of 220 plates.
- Multidrop 384: (Thermo LabSystems): manifold liquid dispensing. This instrument is useful for accurately delivering a fixed amount of liquid rapidly (about 25 seconds) into either 96 or 384 well plates and plating of cells.
- ELx 405 plate washer, Bio-Tek Instruments: well washing, aspiration, dispensing. This instrument would be useful in performing assays which require multi-step aspiration from and dispensing into wells.
- Aquamax DW4: Liquid handling system that integrates four-fluid dispensing and washing for 96, 384, amd 1536 well washing.
Thermo Spinnaker System II
A linear rail based system which is dedicated to high-content screening.
- ImageXpress Confocal (Molecular Devices): A high throughput spinning disk microscope that also doubles as wide field microscope and can be used for phenotypic screens requiring optical sectioning, subcellular localization, mitochondrial morphology evaluation and other demanding applications. We have over 200TB storage capacity integrated and high speed data analysis capabilities as well as custom module editor software to simplify complex analysis workflows.
- Envision: High speed plate reader for 96 or 384 and 1536 well plates in the following modes: UV-VIS absorbance, luminescence, fluorescence polarization, fluorescence, time resolved fluorescence assay as well as alpha screening.
- Cytomat 6001: CO2 plate incubator with a capacity of over 180 plates.
- ELx 406 plate washer, Bio-Tek Instruments: well washing, aspiration, dispensing. This instrument would be useful in performing assays which require multi-step aspiration from and dispensing into wells. It is also equiped with dual syringe dispensers as well as a peristaltic pump for reagent addition.
All devices are integrated under Momentum software for walk-away automation.
NGS Library Preparation System
A HighRes Engineering online system consisting of Bio Star system with Staubli robotic arm and Cellario Scheduler, SteriStore incubator, Wasp plate sealer, plate piercer as well as a Biomek Fxp liquid handler (with integrated thermo cycler). This is an ideal system for RASLseq applications.
Assay Plate Production System
A system consisting of a Fanuc F7, an Echo 555 acoustic dispense system, two STR-240 incubators, a Wasp plate sealer, an X-Peel de-sealer and an Agilent Bravo liquid handler is available for screening plate generation.
Other Available Instruments
- Biosorter (Union Biometrica): The BioSorter® is a continuous flow system capable of analyzing, sorting and dispensing objects ranging in size from 10 to 1,500 µm. It is useful for sorting zebrafish, C.Elegans, organoids, spheroids and other large objects based on either fluorescence or backscatter/size. Moreover, the MSSR has fitted this biosorter with an in house built aerosol management system that enables us to sort live particles into 96 or even 384 well plates and we can define the number of objects and characteristics of the objects on a per well level enabling screening of large biological objects with defined parameters.
- Qbot: Colony picker and arrayer (Genetix). This instrument has a 72 plate capacity and is used in high-throughput sequencing projects as well as for library construction and maintenance. It uses 96 and 384 well plates, omni-trays and is equipped with a video camera for automated colony recognition and blue-white screening.
- Precision 2000 (BioTek): A 12 channel liquid handler (Bio-Tek) which can be used in a sterile bench and addresses 96 and 384 well plates.
- Various Multidrop 384’s: (Thermo LabSystems) This instrument is useful for accurately delivering a fixed amount of liquid rapidly into either 96 or 384 well plates and plating of cells.
- Acumen Explorer (TTP Labtech): This laser scanning cytometer is currently the fastest high content screening system available. It enables cell-by-cell readouts resulting in a flow-cytometer like data structure useful for phenotypic screening. Examples are screens based on translocation events from the nucleus to the cytosol, cytotoxicity screens and even cell cycle based screening is possible.
- ImageXpress (Molecular Devices): A high throughput epifluorescence microscope which can be used for phenotypic screens requiring the highest quality images such as neurite outgrowth, translocation and certain cell toxicity screens. We have a phase contrast option enabling label free phenotypic screening of zebrafish. Using special objectives and analysis modules, even difficult phenotypes such as the of uptake of bacteria by mammalian cells can be quantified. This systen is integrated with an incubator using a custom Denso robotic arm.
- Mantis(Formulatrix): Low dead-volume, non-contact liquid dispenser that can up to six reagents that allows dispensing complex plates and assay development experiments down to 100nL volumes.
Libraries
CRISPR and other New Resources (NEW)
We have curated various arrayed CRISPR resources such as Cas9 constructs which we keep in bacterial and lentiviral format in stock to generate cell lines on demand. We are also actively building a repository of Cas9 carrying cell lines which are clonal and have been characterized in detail. These are available for use in projects using our screening services only. We have also various other reagents such as overexpression systems that allow for the delivery of larger cDNA’s without the need for viral packaging but still yield stable cell lines. For most of our reagents, we have constitutive as well as inducible formats are available for your use. The build-out of targeted libraries for resource is ongoing and we would love to incorporate your feedback!
Human Genome Wide Arrayed CRISPR Library
We are pleased to offer access to a full genome wide arrayed CRISPR library covering the human genome with 65,000 clones, i.e. each gene will have on average 3 constructs, i.e. 6 guides available. Our library is unique in that it contains two guides per plasmid. Individual clones for your targets and subsets are also available for screening.
We also have several human and mouse genome wide collection pooled CRISPR libraries available for screening. These are particularly useful for phenotypes where a complete gene knockout rather than a modulation of the gene can be couples with lethality or reporter gene assays.
All of our libraries are available as individual clones – i.e. we can provide you CRISPR guides, shRNA’s or cDNA’s within a matter of days at a fraction of the cost of an outside vendor. We also offer cell line generation services drawing on our expertise in CRISPR KO, CRISPR-i, CRISPR-a and cDNA genome engineering strategies.
Small Molecule Libraries
Our compounds libraries are split into 4 segments: Pharmacological validation and repurposing libraries (LOPAC, Prestwick and Microsource spectrum, 430 clinical kinase inhibitors annotated with targets), targeted libraries for high value protein classes, lead-like libraries, and diverse libraries and diverse/smart libraries. All of our compounds are at least 90% pure. On average, we can resupply 90%-95% of the hit compounds as powder for follow up. With the exception of the diverse library (UCLA set) which is a pre-plated set, all of our sets are custom sets and are not likely to be found in another screening facility. We have applied extensive filtering against liabilities such as reactive groups, aggregators etc. The drug-likeness and usefulness of our pharmacological validation and re-purposing libraries as well as our custom libraries is well established. We found a historic sucess rate of about 90-95% in delivering tractable hits from our libraries across all targets.
FDA Approved Drug Library
A unique collection of 1,536 high-purity chemical compounds (all off patent) carefully selected for:
- Structural diversity
- Broad spectrum covering several therapeutic areas (from neuropsychiatry to cardiology, immunology, anti-inflammatory, analgesia and more)
- Known safety and bio-availability in humans
LOPAC Collection
A collection of 1,280 pharmacologically-active drug-like molecules for use in the fields of cell signaling & neuroscience. It covers the following target classes and is extremely useful for assay validation:
- Apoptosis
- G Proteins & Cyclic Nucleotides
- Gene Regulation & Expression
- Ion Channels
- Lipid Signaling
- Multi-Drug Resistance
- Neurotransmission
- Phosphorylation
Microsource Spectrum Collection
2000 biologically active and structurally diverse compounds from our libraries of known drugs, experimental bioactives, and pure natural products.
Cysteine Reactive Library (Covalent Inhibitor Library)
3500 drug-like (they are rule of 5 or better) compounds with warheads against cysteines. This library contains a diverse selection of different warheads with specific properties tuned to ensure enough reactivity towards cysteine targets while also maintaining specificity.
Kinase Inhibitor Library
This set includes 2,750 annotated kinase inhibitors covering about more than 1/3rd of the kinome enabling pathway dissection.
GPCR library
This set contains 2290 GPCR modulators that are annotated with targets that cover classes 5-HT Receptor, Dopamine Receptor, Opioid Receptor, Adrenergic Receptors, Cannabinoid Receptor, mGluR, ETA Receptor, etc.
Druggable Compound Set
A set of about 8000 compounds which are targeted at kinases, protease, ion channels and GPCR’s. These compounds are designed to match our siRNA efforts which are based on sets of siRNA’s for the druggable human and mouse genome. This set of 8k compounds was subjected to high-throughput docking to kinases,proteases, ion channels and GPCR’s and included in this set based on their predicted ability to bind to these high-value targets. The drug-likeness of this set is excellent, compounds from this set obey the rule of 5 – many of them even the rule of 4.
Lead Like Compound Set
This set of 20,000 compounds has been custom tailored for us for lead likeness. The compounds in this set are not to be found in any of the traditional “Russian” collections. The lead-like library obeys the more stringent rule of 4 rather than the more permissive rule of 5. This library was selected from a set of about 250,000 compounds to yield compounds which have more favorable properties for subsequent medchem optimization as the average molecule from this set is smaller, has fewer h-acceptor or donor sites and a better logP. It addresses the typical problem that during chemical optimization the compounds typically get heavier and less drug-like leading to more potential for ADME-problems in subsequent stages.
Chemically Diverse Libraries (CombiChem and Traditional Liquid Phase Medicinal Chemistry Libraries)
ChemBridge DiverSet, 30,000 chemically diverse small molecules. A custom set of 20,000 compounds which have been selected for low cellular toxicity and excellent coverage of the chemical space. This diverse library is the DiverSet E from Chembridge – it’s a well-established 30k compound set which contains a lot of interesting structures and has generated many interesting hits in our hands. This set contains a vast structural diversity which has been selected from over 500,000 compounds.
Three smart sets of 20,000, 40,000 and 50,000 compounds each with excellent coverage of the chemical space selected from a set of over 850,000 compounds. Our diverse smart libraries were selected from large compounds sets of 600k and 250k compounds. Using large computer clusters with GPU processors (250 nodes and more) at the CNSI, we filtered the sets for compounds which were drug like and did not have any other liabilities, fewer than 8 rotatable bond etc. and then broke the compounds into clusters of similar structures. This was a challenging process which took about 3 weeks computing time on the cluster. We then sampled these clusters for diversity and included the resulting compounds into our libraries. The resulting compound libraries have excellent properties (at least rule of 5 or better) and they are proprietary to the MSSR.
UCLA In-House Collection
Various professors have donated their compounds to our in-house collection and so far we have collected about 5000 compounds. These libraries are extremely diverse and range from natural products to compounds which have been synthesized using diversity-oriented synthesis.
Compound Properties
Overview of the molecular properties of the smart library space – 300k compounds are available at the MSSR. As of note, the compounds are Lipinski rule of five compliant, with many of them being compliant with the rule of 4 which is a stricter rule set frequently employed in pharmaceutical companies to ensure optimizability during hit-to-lead campaigns. Of particular note is that our compound sets are heavily enriched in SP3 content, which was design feature during compound selection. We have curated our compound libraries specifically with 3D-dimensionality in mind in order to have the best change of finding compounds that can bind into a multitude of targets. Moreover, most of our compounds have a CNS MPO score >4, which makes them also suitable for CNS drug discovery projects and decreases the likelihood of being a drug efflux pump substrate which is extremely important for novel cancer therapeutics. Through historical data analysis, we have seen that our compound deck provides tractable chemical matter in ~90% of our projects.
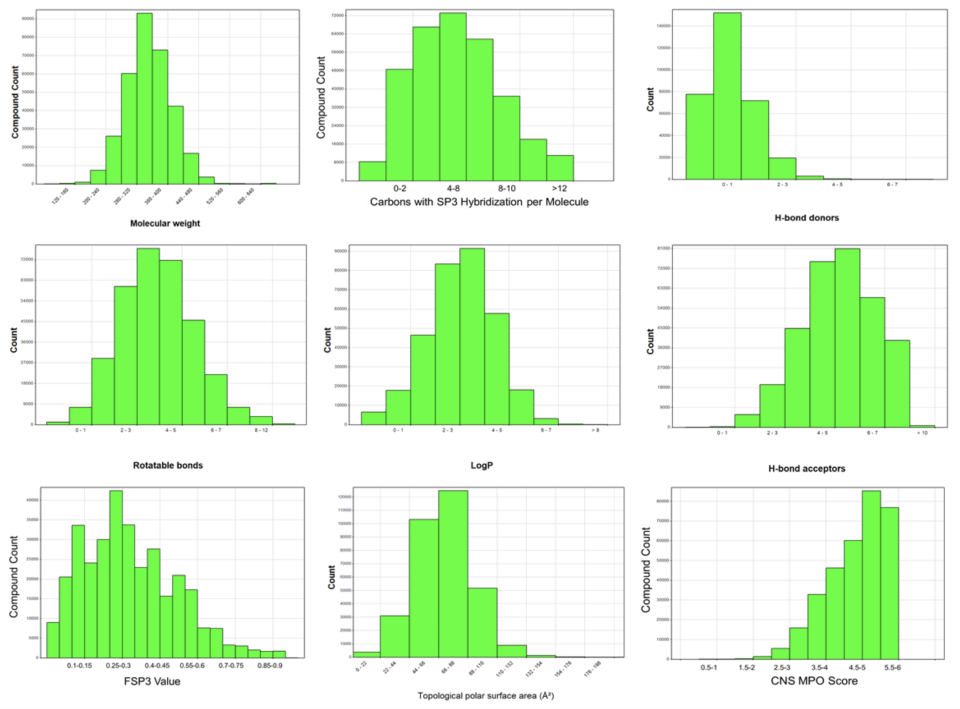
Arrayed siRNA Libraries
A human and mouse druggable genome wide collection is available for screening. The two libraries cover about 7,000 genes each. We have four siRNA’s per target, which are screened in a non-pooled format. Kinases and GPCR’s are available as sub-libraries.
Arrayed shRNA Library
Our arrayed shRNA libraries contain over 60,000 clones targeting about 18,000 genes with an emphasis in coverage on desirable target classes such as kinases, proteases, phosphatases, GPCRs and ion channels. In addition to the traditional target classes we offer various custom libraries such as a validated cancer specific shRNA set of about 150 targets from the NCI and a cancer specific set of over 2000 clones covering 500 cancer related genes selected by the Sanger Institute. Other sublibraries cover transcription factors, ubiquitinization and de-ubiquitinization pathways, apoptosis etc. These libraries are proprietary to the MSSR as they were custom selected and arrayed at the MSSR. The capability of customizing shRNA libraries enables the MSSR as well to tailor shRNA libraries to the needs of a particular Principle Investigator’s laboratory.
Arrayed cDNA Library
A human and mouse mixed genome wide collection is available for screening. The libraries covers about 16,000 genes and is a consolidated version of the expressible clones from the MGC collection of full length cDNA clones. These clones are in the pSPORT6 expression vector which uses the CMV promoter to drive the expression of the insert. We can as well customize libraries to your requirements as needed.
We also offer a 13,000 clones strong lentiviral cDNA library in a pLX304 vector with V5 tag on the c-terminus and also blasticidin resistance for selection.
Yeast Knockout Collection
A genome wide set of non-essential knockout mutants of yeast is available for screening in both mating types (alpha and A). The strain collections have been condensed into 384 well plates to facilitate high throughput screening. We have successfully used this library as well for synthetic genetic array screens.
e.coli Knockout Collection
A genome wide set of non-essential knockout mutants of E-Coli is available for screening. The strain collections have been condensed into 384 well plates to facilitate high throughput.
e. coli Promoter Collection
A set of about about 2000 E-Coli promoters with GFP as reporter gene is available for screening. The strain collections have been condensed into 384 well plates to facilitate high throughput screening.
Pricing
There is a tiered fee structure. Costs for Cancer Center members are listed below; for non-members, please refer to our website at www.mssr.ucla.edu for details.
| SERVICE | JCCC RATE |
|---|---|
| Assay development | $630.60 |
| Small molecule screening (up to 100K) | $549.16 |
| Small molecule screening (additional 100K) | $772.29 |
| shRNA screening (human, per sub-library) | $606.36 |
| siRNA screening (human genome-wide) | $606.36 |
| siRNA screening (mouse genome-wide) | $606.31 |
| Full genome cDNA screening | $575.53 |
| Access to clones (cDNA) | $12.00 |
| Full technical support (per hour) | $87.37 |
| Data analysis (per hour) | $30.17 |
