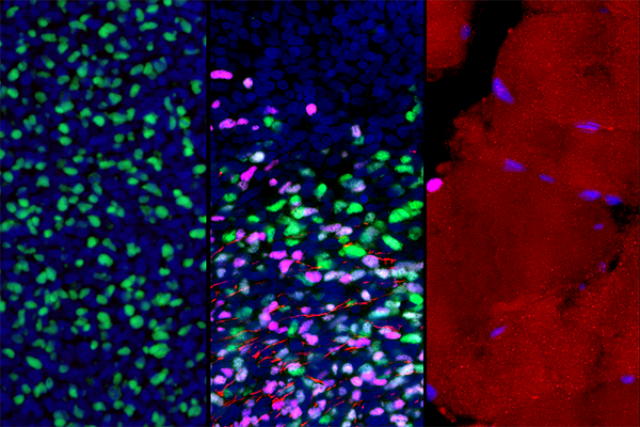
An interdisciplinary team of researchers at the Eli and Edythe Broad Center for Regenerative Medicine and Stem Cell Research at UCLA has developed a first-of-its-kind roadmap of how human skeletal muscle develops, including the formation of muscle stem cells.
The study, published May 11 in the peer-reviewed journal Cell Stem Cell, identified various cell types present in skeletal muscle tissues, from early embryonic development all the way to adulthood. Focusing on muscle progenitor cells, which contribute to muscle formation before birth, and muscle stem cells, which contribute to muscle formation after birth and to regeneration from injury throughout life, the group mapped out how the cells' gene networks — which genes are active and inactive — change as the cells mature.
The roadmap is critical for researchers who aim to develop muscle stem cells in the lab that can be used in regenerative cell therapies for devastating muscle diseases, including muscular dystrophies, and sarcopenia, the age-related loss of muscle mass and strength.
"Muscle loss due to aging or disease is often the result of dysfunctional muscle stem cells," said April Pyle, senior author of the paper and a member of the Broad Stem Cell Research Center. "This map identifies the precise gene networks present in muscle progenitor and stem cells across development, which is essential to developing methods to generate these cells in a dish to treat muscle disorders."
Researchers in Pyle's lab and others around the world already have the capacity to generate skeletal muscle cells from human pluripotent stem cells — cells that have the ability to self-renew and to develop into any cell type in the body. However, until now, they had no way of determining where these cells fall on the continuum of human development.
"We knew that the muscle cells we were making in the lab were not as functional as the fully matured muscle stem cells found in humans," said Haibin Xi, first author of the new paper and an assistant project scientist in Pyle's lab. "So we set out to generate this map as a reference that our lab and others can use to compare the genetic signatures of the cells we are creating to those of real human skeletal muscle tissue."
To create this resource, the group gathered highly specific data about two different groups of skeletal muscle cells: those from the human body, ranging from the fifth week of embryonic development to middle age, and those derived from human pluripotent stem cells the researchers generated in the lab. They then compared the genetic signatures of cells from both sources.
The group obtained 21 samples of human skeletal muscle tissue from their UCLA collaborators and from colleagues at the University of Southern California and the University of Tübingen in Germany. For the pluripotent stem cell–derived muscle cells, the group evaluated cells created using their own unique method and the methods of several other groups.
The Pyle lab collaborated with the lab of Kathrin Plath, a UCLA professor of biological chemistry and member of the Broad Stem Cell Research Center, to conduct high-throughput droplet-based single-cell RNA sequencing of all of the samples. This technology enables researchers to identify the gene networks present in a single cell and can process thousands of cells at the same time. Leveraging the power of this technology and the Plath lab's bioinformatics expertise, the group identified the genetic signatures of various cell types from human tissues and pluripotent stem cells.
They next developed computational methods to focus on muscle progenitor and stem cells and mapped out their gene networks associated with every developmental stage. This enabled the group to match the genetic signatures found in the pluripotent stem cell–derived muscle cells with their corresponding locations on the map of human muscle development.
The group found that pluripotent stem cell–derived muscle cells produced by all the methods they tried resembled muscle progenitor cells at an early developmental state and did not align to adult muscle stem cells.
In addition to pinning down the true maturity of the lab-produced cells, this analysis also provided details about the other cell types present in skeletal muscle tissue across development and in populations derived from human pluripotent stem cells. These cells could play an essential role in muscle cell maturation and could be critical to improving methods to generate and support muscle stem cells in a dish.
"We found that some methods to generate muscle cells in a dish also produce unique cell types that likely support the muscle cells," said Pyle, who is also a member of the UCLA Jonsson Comprehensive Cancer Center. "And so now our questions are, what are these cells doing? Could they be the key to producing and supporting mature and functional muscle stem cells in a dish?"
Moving forward, Pyle and her colleagues will focus on harnessing this new resource to develop better methods for generating muscle stem cells from human pluripotent stem cells in the lab. She hopes that by focusing on the stem cell–associated gene expression networks and supportive cell types they identified, they can produce high-powered muscle stem cells that can be useful for future regenerative therapies.
This research was supported by the California Institute for Regenerative Medicine; the National Institutes of Health; a UCLA Broad Stem Cell Research Center Rose Hills Foundation Innovator Grant; the David Geffen School of Medicine at UCLA; the UCLA Jonsson Comprehensive Cancer Center and UCLA Broad Stem Cell Research Center Ablon Scholars Program; the Howard Hughes Medical Institute; a UCLA Broad Stem Cell Research Center Rose Hills Foundation Graduate Scholarship; and the UCLA Tumor Cell Biology Training Program.