An interdisciplinary team of scientists from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA has received a $4.7 million award from the National Institutes of Health (NIH) to study how genomic variation influences the production of induced pluripotent stem cells.
The five-year grant to UCLA researchers Brunilda Balliu, Jason Ernst, Chongyuan Luo, Kathrin Plath, Roy Wollman and Noah Zaitlen is one of 25 grants the NIH’s National Human Genome Research Institute announced today as part of its newly established Impact of Genomic Variation on Function (IGVF) Consortium.
The term genomic variation refers to the differences in the sequence of DNA from one person to another. Humans have genomic sequences that are 99.9% identical, but the 0.1% that varies from individual to individual – their genomic variants – help determine each person’s physical characteristics and disease risk. The IGVF Consortium aims to identify how genomic variation changes human genome function, and how that, in turn, influences human health and disease.
The interdisciplinary UCLA team will use their IGVF grant to create an open-access induced pluripotent stem cell bank and genomic data library intended to make the process of developing stem cell therapies more efficient.
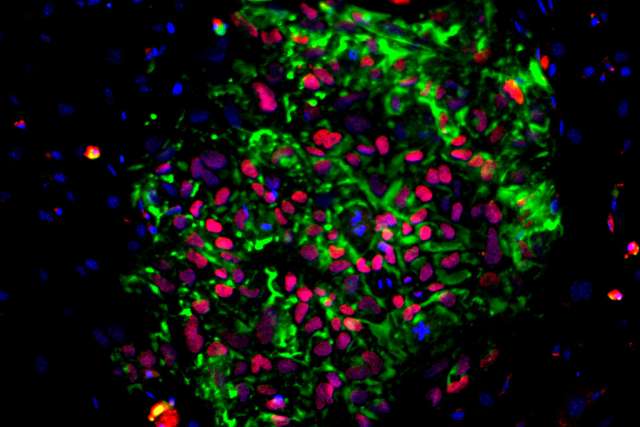
Human induced pluripotent stem cells, or iPS cells, are derived from tissue-specific cells such as skin or blood cells by reprogramming, a method that converts the cells back to a stem-like state. IPS cells retain the genomic characteristics of the person from whom they were created, can make unlimited copies of themselves and can be coaxed to differentiate, or give rise to any cell type found in the body. These qualities make iPS cells an ideal model system for studying human diseases and an attractive potential source for cell therapies for a range of diseases and injuries.
“Induced pluripotent stem cells are the foundation of regenerative medicine, but before we can realize their full potential, we need to understand them more deeply,” said Chongyuan Luo, leader of the project and assistant professor of human genetics at the David Geffen School of Medicine at UCLA. “We know that the tissue-specific cells we obtain from individual donors have unique genetic characteristics that affect how they respond to the techniques we use to direct reprogramming and differentiation, but we don’t understand why.”
To begin to solve this mystery, Luo and his collaborators will study iPS cells derived from 100 healthy individuals from a range of ethnic and racial backgrounds using whole genome sequencing and several other cutting-edge technologies. They will then use computational modeling to connect the genomic variations present in each of these iPS cells with the differences in their reprogramming and differentiation capacities.
In keeping with the IGVF’s open access model, the cells used in this study and the data uncovered about them will be made freely available to the global research community. The UCLA team envisions the outcome of their project will be an iPS cell library from which scientists can obtain iPS cells along with troves of data about their genomic variations and how those variations influence their behavior and potential.
In addition to sharing this resource with the scientific community, the team aims to use the insights gained from this project to develop a universal reprogramming method that will allow for efficient iPS cell derivation and differentiation regardless of genomic variants.
“Ultimately, we hope the approach we develop will pave the way for next-generation therapies that will be more efficient and therefore more affordable and accessible for patients,” said Plath, a collaborator on the project and professor of biological chemistry