Microbiome
Neuroimaging Core
We provide a wide range of services to support researchers and clinicians interested in multimodal neuroimaging data obtained from human studies related to existing data sets, ongoing studies, new studies and grant development, to name a few.
Our automated pipelines are able to handle processing from using various acquisition protocols.
For more information, contact Arpana Church, PhD, Core Director, at [email protected]
The major categories of services provided by the Neuroimaging Core
- Consultation on data acquisition and experimental design
- Brain preprocessing of raw brain images
- Quality control
- Preprocessing
- Neuroimaging data will undergo quality control using our optimized and standardized Brain Imaging Data Structure (BIDS)-App, MRIQC and established protocols. BIDS-Apps have been developed to assess a wide range of quality control metrics, and implement a growing number of popular functional and structural MRI analyses
- Advanced neuroimaging methodologies including
- Regional brain grey matter morphometry (structural magnetic resonance imaging),
- Functional (resting state and task-based fMRI)
- Anatomical microstructural connectivity (diffusion spectrum imaging)
-
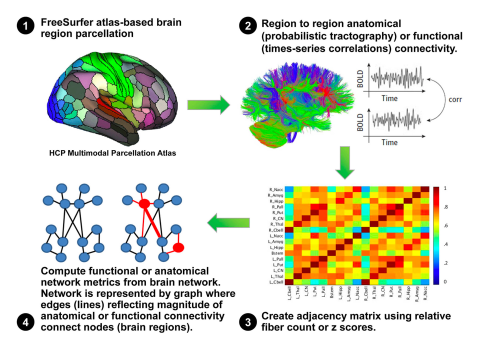
Advanced multimodal data analyses
Novel approaches for large datasets such as network-based analysis (e.g., Graph theory-based association network analysis). See computational pipeline (on right.)
- Data management and storage
- Assistance with interpretation of analysis results
- Research project development for manuscripts and grants
- Coordination with the other cores of the Microbiome Center (e.g., Data Core, Microbiome Core and Bioinformatics and Biostatistics Core)
Facilities and equipment
- The Neuroimaging Core (NIC) is housed in the renovated 6,000 sq ft space in the UCLA Center for Health Sciences, 42-210. The key personnel for the proposed NIC have their offices in this facility.
- Software required for state-of-the-art Magnetic Resonance Imaging (MRI) analysis techniques: grey matter parcellation (Freesurfer), anatomical connectivity (diffusion tensor Imaging; Camino software), functional connectivity (resting state imaging; CONN software), graph theory based complex network analysis, connectivity gradient analysis, multivariate analysis.
- Established international imaging analysis programs that have been extended by custom scripting and internally developed algorithms. The library of available analysis programs provides all major mathematical, neuroimaging, and statistical packages, including AFNI, Matlab, R, SPM, FSL, Freesurfer, and many others.
- Utilize an integrated system of 112 CPU cores, 188 GB of RAM, and 80+ TB of cloud-hosted storage with automated nightly backups.
- System consists of three virtual environments (VM), one CPU-focused, one GPU-focused, and one database-focused. All run on Ubuntu 20.04 and are capable of being upgraded or scaled as needed. The environments are maintained and managed by UCLA’s IT team (DGIT). All data is automatically synchronized between each of the VMs while still allowing for separate storage within each VM, thus accommodating multiple performance levels to meet the needs for any given task. The CPU-focused VM consists of 64 CPU cores and 126 GB of RAM. The GPU-focused VM has 48 CPU cores and 62 GB of RAM, and the GPU allows for parallelization / hardware acceleration of certain tasks. The two VMs together total over 100 compute nodes. The database-focused VM is separate to ensure no resources are taken from computing jobs.
- Data transfer across servers via a gigabit storage area network (SAN).
- Additional storage resides on campus grid systems (grid.ucla.edu) and the new UCLA cloud service (cloud.ucla.edu).
- No access to central repository data is available outside the UCLA and CNSR firewalls.
- Collaboration via internet-accessible data stores occurs in a monitored, restricted network DMZ.